基本情况
焦文标,华中农业大学信息学院教授、博士生导师,果蔬园艺作物种质创新与利用全国重点实验室、湖北洪山实验室、农业生物信息湖北省重点实验室固定研究人员,先后获国家级、省级等人才项目资助。
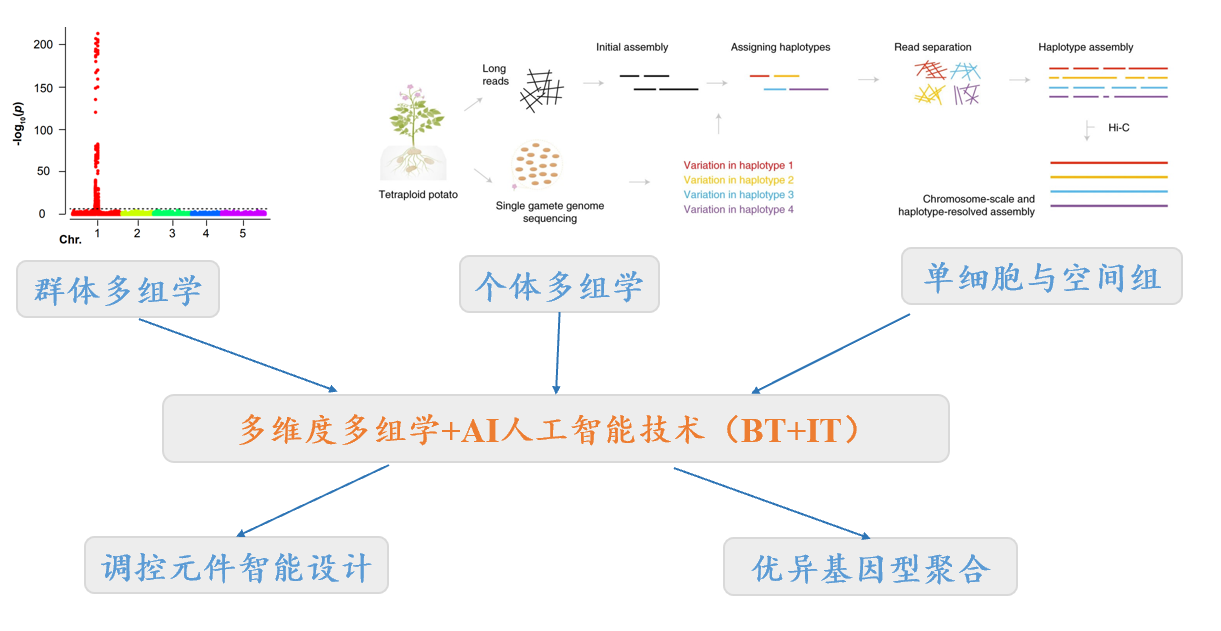
本课题组依托果蔬园艺作物种质创新与利用全国重点实验室、湖北洪山实验室等,以马铃薯、柑橘等园艺作物为主要研究对象,基于高通量组学技术(如三代测序、单细胞组学、时空组学等技术)和深度学习等人工智能技术,开发生物信息学算法,以解决基因组组装、遗传变异与泛基因组、基因组进化、共性调控元件挖掘与智能设计等领域的关键问题;为园艺作物的遗传改良和品质创新提供理论指导。本课题组主要研究成果发表在Nature Genetics, Genome Biology, Genome Research, Nature Communications, Molecular Biology and Evolution等期刊上。
欢迎有生物信息学、生物学、计算机科学、统计学等相关学科背景的本科生、硕士/博士研究生、博士后等加入我们课题组。本课题组长年招聘博士后,欢迎邮件联系。
主要研究兴趣:
基因组组装、遗传变异与泛基因组等算法开发与应用 Development and application of algorithms for genome assembly and graph pangenomes
1)Genome Assembly 针对杂合二倍体、同源多倍体等复杂基因组,开发单倍型组装、Telomere-to-Telomere(T2T)组装算法,以获得高质量的柑橘、马铃薯等杂合二倍体或多倍体的完整单倍型序列。(Jiao et al., Genome Res. 2017; Sun et al., Nat. Genet. 2022; Xiao et a., Horti. Res. 2023)
2) Graph Pangenome 基于图形泛基因组理论,开发快速、高效、经济的遗传变异分型算法,解决当前算法对植物复杂变异分型效果较差的难题,并应用于柑橘、马铃薯等杂合二倍体和多倍体园艺作物,以及玉米、小麦等大型基因组的各类遗传变异(SNP、Indel、SV)的分型。(Du et., Genome Biology 2024; Du et al., Molecular Plant 2025; Du et al., aBiotech 2025)。
3)Population Genomics 通过大规模鉴定植物基因组遗传变异,开发比较或进化基因组学算法(例如:共线性多态性Syntenty diversity、跨物种超级泛基因组),解析基因组进化机制和遗传变异功能效应(如遗传不亲和) (Jiao et al., Nat. Communi. 2021;Jiao et., Mol. Biol. Evol. 2022; Du et al., Plant Communications 2024)。
园艺作物多组学大数据与智能设计育种 Multi-omic bigdata and AI-guided breeding for horticultural crops
1) 马铃薯多组学与智能设计育种:通过组学算法的创新,如多倍体基因组单倍型组装、多倍体遗传变异识别与分型、多倍体GWAS & GS、多倍体基因组深度学习模型等,助力多倍体马铃薯种质资源基因型鉴定、功能基因挖掘、基因组智能设计育种等。(Lian et al., Molecular Plant 2025)
2) 园艺作物共性调控元件的挖掘与应用:综合利用转录组、调控组、单细胞和时空组学等多组学技术,挖掘园艺作物共性性状的调控元件和网络;开发深度学习模型,智能设计与优化调控元件。
3) 园艺大数据平台构建与维护:通过与本室其他课题组的合作,共同构建园艺作物多组学大数据整合分析与研究平台。
教育经历
• 2014.01-2017.06,德国马克斯普朗克植物育种研究所,生物信息学,博士
• 2014.01–2018.01,德国科隆大学(联合培养),生物信息学,博士
• 2010.09–2013.06,华中农业大学,生物化学与分子生物学,硕士
• 2006.09–2010.06,华中农业大学,生物学基地班,学士
工作经历
• 2021.04-至今,华中农业大学信息学院 教授
• 2021.01-2021.03 德国慕尼黑大学 博士后
• 2017.07-2020.12 德国马克斯普朗克植物育种研究所 博士后
科研项目
(1)华中农业大学高层次人才启动经费 在研 主持 (2021.4-2026.4)
(2)国家青年人才项目 在研 主持 (2023.1-2025.12)
(3)国家自然科学基金面上项目 在研 主持 (2023.1-2026.12)
(4)武汉英才优秀青年人才 在研 主持 (2023.1-2024.12)
(5)重点实验室自主培育重点项目 在研 主持 (2023.9-2028.8)
(6) 国家重点研发计划-政府间国家创新合作重点专项 项目骨干 (2024.6-2027.5)
(7)西藏自治区科学技术厅重点研发专项 项目骨干 (2025.1-2026.12)
主讲课程
本科生课程
《生物信息学原理》
《生物信息软件综合实践》
《写作与沟通:人工智能与生命科学》
主要研究论文:ORCiD http://orcid.org/0000-0001-8355-2959
一作(含共同一作#) 或通讯作者(含共同通讯作者*)文章
15. Lian Q, Jiao W-B, Wang Y: Designing Better Crops with Phased Pangenomes. Mol Plant 2025, doi:10.1016/j.molp.2025.08.014.
14. Du Z-Z, He J-B, Xiao P-X, Hu J, Yang N, Jiao W-B: Varigraph: An accurate and widely applicable pangenome graph-based variant genotyper for diploid and polyploid genomes. Mol Plant 2025, 18:1587–1601.
13.Du Z-Z, He J-B, Jiao W-B: Plant graph-based pangenomics: techniques, applications, and challenges. aBIOTECH 2025, doi:10.1007/s42994-025-00206-7.
12. Wang C, Tan L, Zhang Z, Li X, Xia L, Cao P, Tong H, Ou X, Li S, Zhang J,Li et al.: Haplotype-resolved genome reveals haplotypic variation and the biosynthesis of medicinal ingredients in Areca catechu L. Mol Hortic 2025, 5:24.
11. Zuo H, Liu S, Tan L, Huang Y, Li Y, Gesang P, Hong Y, Deng X, Wang X, Xu Q, et al.: Chromosome-level assembly of Prunus serrula Franch genome. Sci Data 2025, 12:494.
10. Du, Z.-Z., He, J.-B., and Jiao, W.-B*. (2024). SynDiv: An efficient tool for chromosome collinearity-based population genomics analyses. Plant Commun. 2024, 5:101071. doi:https://doi.org/10.1016/j.xplc.2024.101071.
9.Du Z-Z, He J-B, Jiao W-B*: A comprehensive benchmark of graph-based genetic variant genotyping algorithms on plant genomes for creating an accurate ensemble pipeline. Genome Biology 2024, 25:91 https://doi.org/10.1186/s13059‑024‑03239‑1
8. Song L, Huang Y, Zuo H, Tang N, Li Z, Jiao WB*, Xu F*, Xu Q*, Chen Z*: Chromosome-level assembly of triploid genome of sichuan pepper (Zanthoxylum armatum). Hortic Plant J 2024, 10:437–449.
7. Xiao P-X, Li Y, Lu J, Zuo H, Pingcuo G, Ying H, Zhao F, Xu Q, Zeng X, Jiao W-B*: High-quality assembly and methylome of a Tibetan wild tree peony genome ( Paeonia ludlowii) reveal the evolution of giant genome architecture. Hortic Res 2023, 10.
6. Sun H#, Jiao WB#, Krause K, Campoy JA, Goel M, Folz-Donahue K, Kukat C, Huettel B, Schneeberger K: Chromosome-scale and haplotype-resolved genome assembly of a tetraploid potato cultivar. Nature Genetics 2022, 3:342-348
5. Jiao WB, Patel V, Klasen J, Liu F, Pecinkova P, Ferrand M, et al. :The Evolutionary Dynamics of Genetic Incompatibilities Introduced by Duplicated Genes in Arabidopsis thaliana. Molecular Biology and Evolution 2021, 38:1225-1240
4. Jiao WB, Schneeberger K: Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nature Communications 2020, 11:989.
3. Jiao WB, Schneeberger K: The impact of third generation genomic technologies on plant genome assembly. Current Opinion Plant Biology 2017, 36:64-70.
2. Jiao WB, Garcia Accinelli G, Hartwig B, Kiefer C, Baker D, Severing E, Willing E-M, Piednoel M, Woetzel S, Madrid-Herrero E, et al.: Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Research 2017, 27:778–786.
1. Jiao WB, Huang D, Xing F, Hu Y, Deng XX, Xu Q, Chen LL: Genome-wide characterization and expression analysis of genetic variants in sweet orange. Plant Journal 2013, 75:954–964.
合作参与文章
19. Fan Y-J, Du Z-Z, He X-Y, Liu Z-A, Zhuang J-X, Xiao G-A, Duan Y-Y, Tan F-Q, Xie K-D, Jiao W-B, et al.: Somatic variations in the meiosis-specific gene CrMER3 confer seedlessness in a citrus bud sport. J Integr Plant Biol 2025, n/a.
18. Liu S, Xu Y, Yang K, Huang Y, Lu Z, Chen S, Gao X, Xiao G, Chen P, Zeng X, et al.: Origin and de novo domestication of sweet orange. Nat Genet 2025, 57:754–762.
17. Hu S, Zeng X, Liu Y, Li Y, Qu M, Jiao W-B, Han Y, Kang C: Global characterization of somatic mutations and DNA methylation changes during vegetative propagation in strawberries. Genome Res 2024, 34:1582–1594.
16. Sun H, Abeli P, Campoy JA, Rütjes T, Krause K, Jiao W-B, Beaudry R, Schneeberger K: The identification and analysis of meristematic mutations within the apple tree that developed the RubyMac sport mutation. BMC Plant Biol 2024, 24:912.
15. Del Toro-De León, G., van Boven, J., Santos-González, J., Jiao, W.-B., Peng, H., Schneeberger, K., and Köhler, C. (2024). Epigenetic and transcriptional consequences in the endosperm of chemically induced transposon mobilization in Arabidopsis. Nucleic Acids Res. Advance Access published July 5, 2024, doi:10.1093/nar/gkae572.
14. Hu J, Guo F, Du Z, Chen P, Shi C, Zhang J, Ye J, Deng X, Larkin RM, Jiao W, et al.: A rare inter-haplotypic recombination at the S-locus contributed to the loss of self-incompatibility in trifoliate orange. Plant Commun 2024, doi:10.1016/j.xplc.2024.100940.
13. Yuan-yuan LIU, Jian-ke D, Jing-wen Y, Wen-xiang MEI, Gang C, Jing-jing GUO: 利用野生种 Solanum boliviense 创制马铃薯抗寒种质. 作物学报 2024, doi:10.3724/SP.J.1006.2024.34159.
12. 宋波涛, 涂卫, 王海波, 董建科, 袁平平, 景晟林, 刘园园, 焦文标: 马铃薯种质演化与分类研究进展. 中国马铃薯 2023, 3:536–554.
11. Huang Y, He J, Xu Y, Zheng W, Wang S, Chen P, Zeng B, Yang S, Jiang X, Liu Z, et al.: Pangenome analysis provides insight into the evolution of the orange subfamily and a key gene for citric acid accumulation in citrus fruits. Nat Genet 2023, 55:1964–1975.
10. Madrid E, Severing E, de Ansorena E, Kiefer C, Brand L, Martinez-Gallegos R, Woetzel S, Kemi U, Jiao WB, Schneeberger K, et al.: Transposition and duplication of MADS-domain transcription factor genes in annual and perennial Arabis species modulates flowering. Proc. Natl. Acad. Sci. 2021, 118.
9. Campoy, J.A., Sun, H., Goel, M. Jiao WB, et al. Gamete binning: chromosome-level and haplotype- resolved genome assembly enabled by high-throughput single-cell sequencing of gamete genomes. Genome Biol. 21, 306 (2020).
8. Goel M, Sun H, Jiao WB, Schneeberger K: SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 2019, 20:277.
7. Dittberner H, Becker C, Jiao WB, Schneeberger K, Hölzel N, Tellier A, Meaux J: Strengths and potential pitfalls of hay transfer for ecological restoration revealed by RAD‐seq analysis in floodplain Arabis species. Mol. Ecol. 2019, 28:3887–3901.
6. Kiefer C, Willing E-M, Jiao WB, Sun H, Piednoël M, Hümann U, Hartwig B, Koch MA, Schneeberger K: Interspecies association mapping links reduced CG to TG substitution rates to the loss of gene-body methylation. Nature Plants 2019, 5:846–855.
5. Zapata L, Ding J, Willing E, Hartwig B, Bezdan D, Jiao WB, Patel V, et al.: Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc. Natl. Acad. Sci. 2016, 113:E4052–E4060.
4. Zhang J, Chen LL, Sun S, Kudrna D, Copetti D, Li W, Mu T, Jiao WB, Xing F, Lee S, et al.: Building two indica rice reference genomes with PacBio long-read and Illumina paired-end sequencing data. Sci. Data 2016, 3:160076.
3. Zhang J, Chen LL, Xing F, Kudrna DA, Yao W, Copetti D, Mu T, Li W, Song JM, Xie W, Lee S, Talag J, Shao L, An Y, Zhang CL, Ouyang Y, Sun S, Jiao WB, et al.: Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minghui 63. Proc. Natl. Acad. Sci. 2016, 113:E5163–E5171.
2. Gutjahr C, Gobbato E, Choi J, Riemann M, Johnston MG, Summers W, Carbonnel S, Mansfield C, Yang SY, Nadal M, Acosta I, Takano M, Jiao WB, et al.: Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 2015, 350:1521–1524.
1. Xu Q, Chen LL, Ruan X, Chen D, Zhu A, Chen C, Bertrand D, Jiao WB, Hao BH, Lyon MP, et al.: The draft genome of sweet orange (Citrus sinensis). Nature Genetics 2013, 45:59–66.
